Identify biomarkers
find_biomarker(
MAE,
tax_level,
input_select_target_biomarker,
nfolds = 3,
nrepeats = 3,
seed = 99,
percent_top_biomarker = 0.2,
model_name = c("logistic regression", "random forest")
)Arguments
- MAE
A multi-assay experiment object
- tax_level
The taxon level used for organisms
- input_select_target_biomarker
Which condition is the target condition
- nfolds
number of splits in CV
- nrepeats
number of CVs with different random splits
- seed
for repeatable research
- percent_top_biomarker
Top importance percentage to pick biomarker
- model_name
one of 'logistic regression', 'random forest'
Value
A list
Examples
data_dir <- system.file("extdata/MAE.rds", package = "animalcules")
toy_data <- readRDS(data_dir)
p <- find_biomarker(toy_data,
tax_level = "family",
input_select_target_biomarker = c("DISEASE"),
nfolds = 3,
nrepeats = 3,
seed = 99,
percent_top_biomarker = 0.2,
model_name = "logistic regression"
)
#> Loading required package: ggplot2
#> Loading required package: lattice
p
#> $biomarker
#> biomarker_list
#> 1 Carnobacteriaceae
#> 2 Coccodiniaceae
#> 3 Comamonadaceae
#> 4 Didymellaceae
#> 5 Malasseziaceae
#> 6 Microviridae
#> 7 Oxalobacteraceae
#> 8 Papillomaviridae
#> 9 Streptomycetaceae
#>
#> $importance_plot
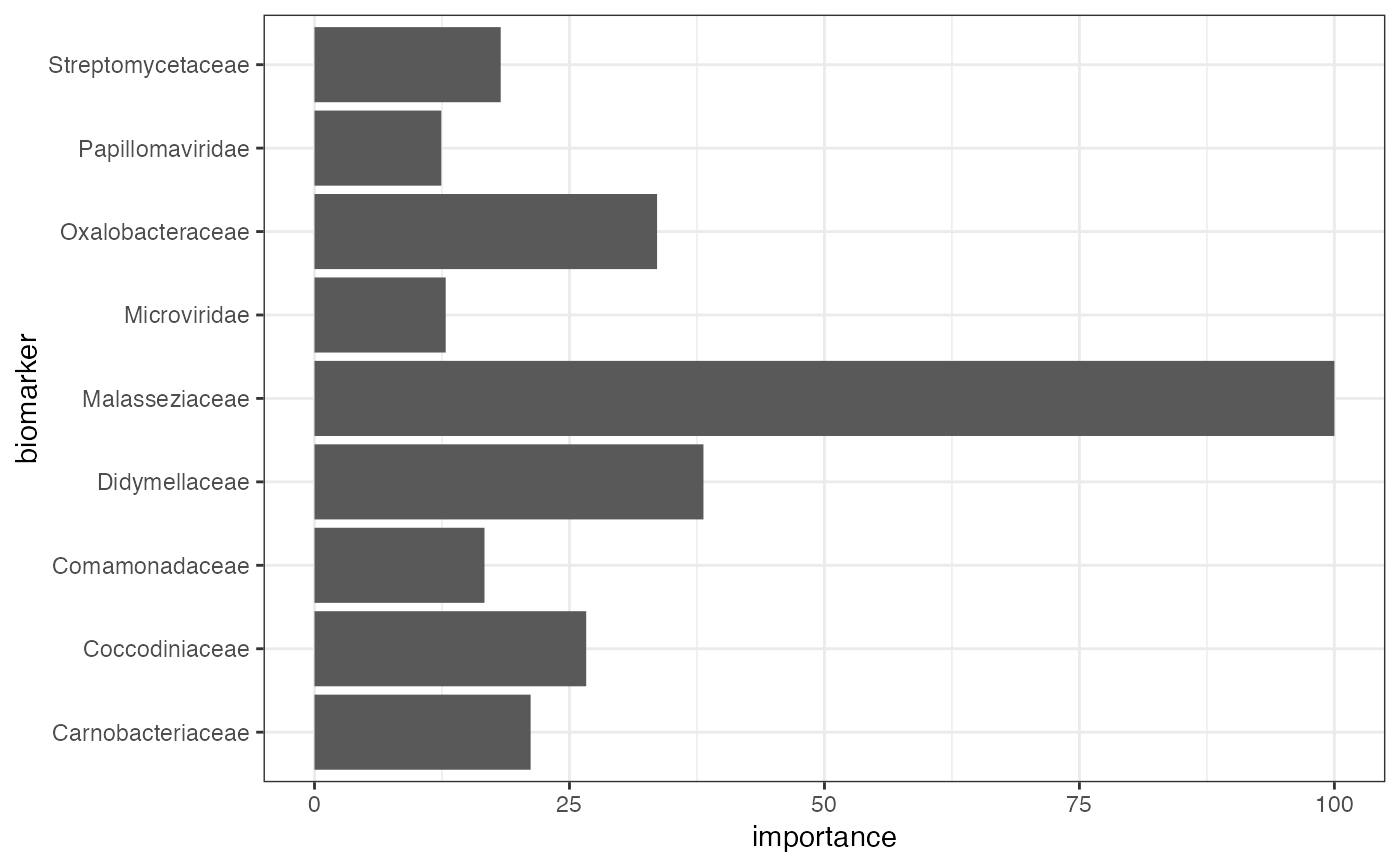 #>
#> $roc_plot
#>
#> $roc_plot
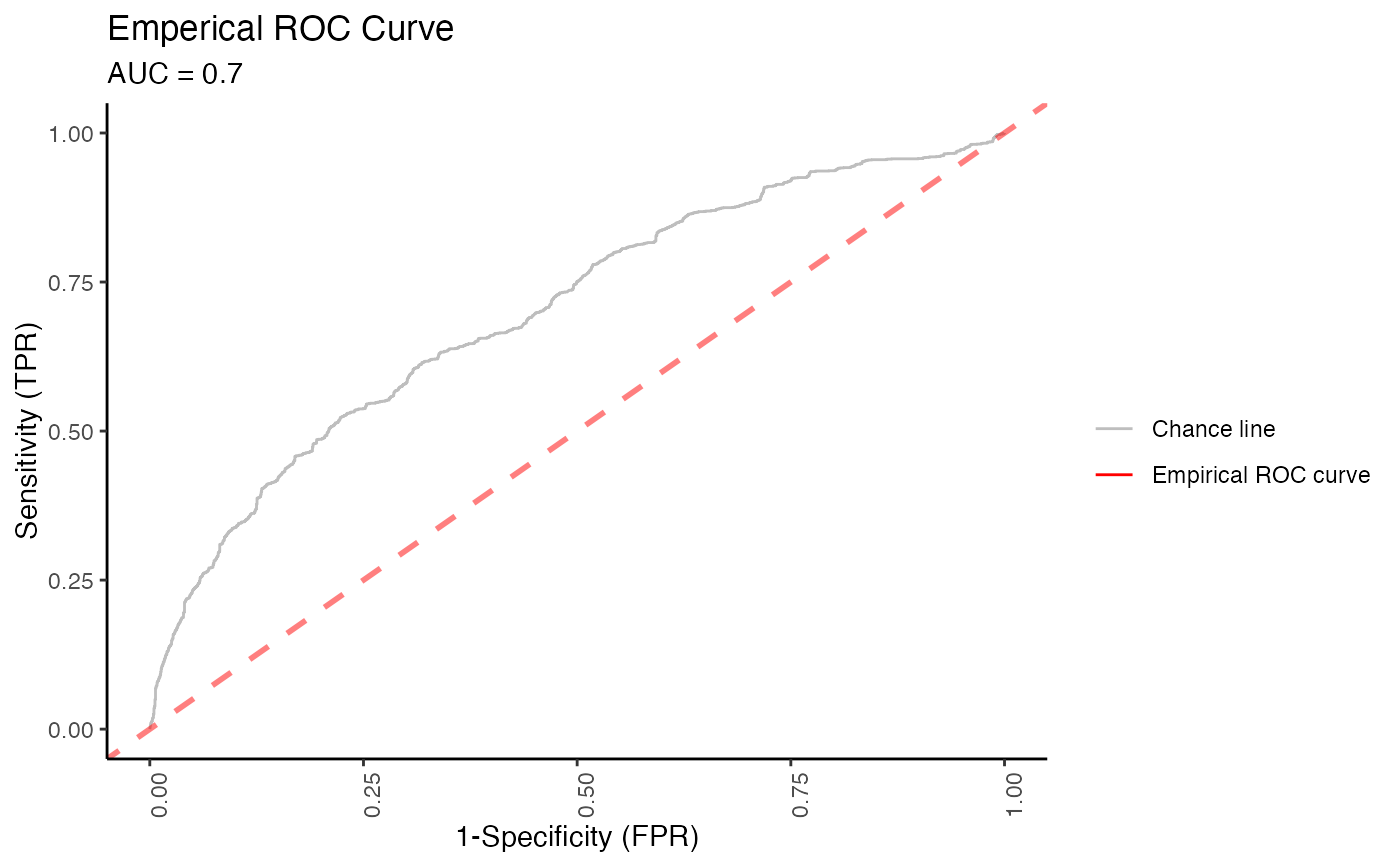 #>
#>