An R-based implementation of the NMIT, a multivariate distance-based test for
group comparisons of microbial temporal interdependence. The NMIT test
provides a comprehensive way to evaluate the association between key
phenotypic variables and microbial interdependence. This function is
recommended for use after a filtering step using
filter_MAE. Note, the "ComplexHeatmap" package is required to
use the plotting features of the function. The function requires the
"vegan" package.
Usage
NMIT(
dat,
unit_var,
fixed_cov,
covariate_time,
method = "kendall",
dist_type = "F",
heatmap = TRUE,
classify = FALSE,
fill_na = 0,
...
)Arguments
- dat
A
MultiAssayExperimentobject specially formatted as an animalcules output.- unit_var
a numeric vector of subject.
- fixed_cov
A character vector of the names of covariates of interest found in
dat.- covariate_time
Character string giving the name of the discrete time-based covariate in the metadata to group abundances by.
- method
an option of the correlation method ("pearson", "kendall", "spearman"). The default method is "kendall".
- dist_type
A character string specifying the type of matrix norm to be computed. The default is
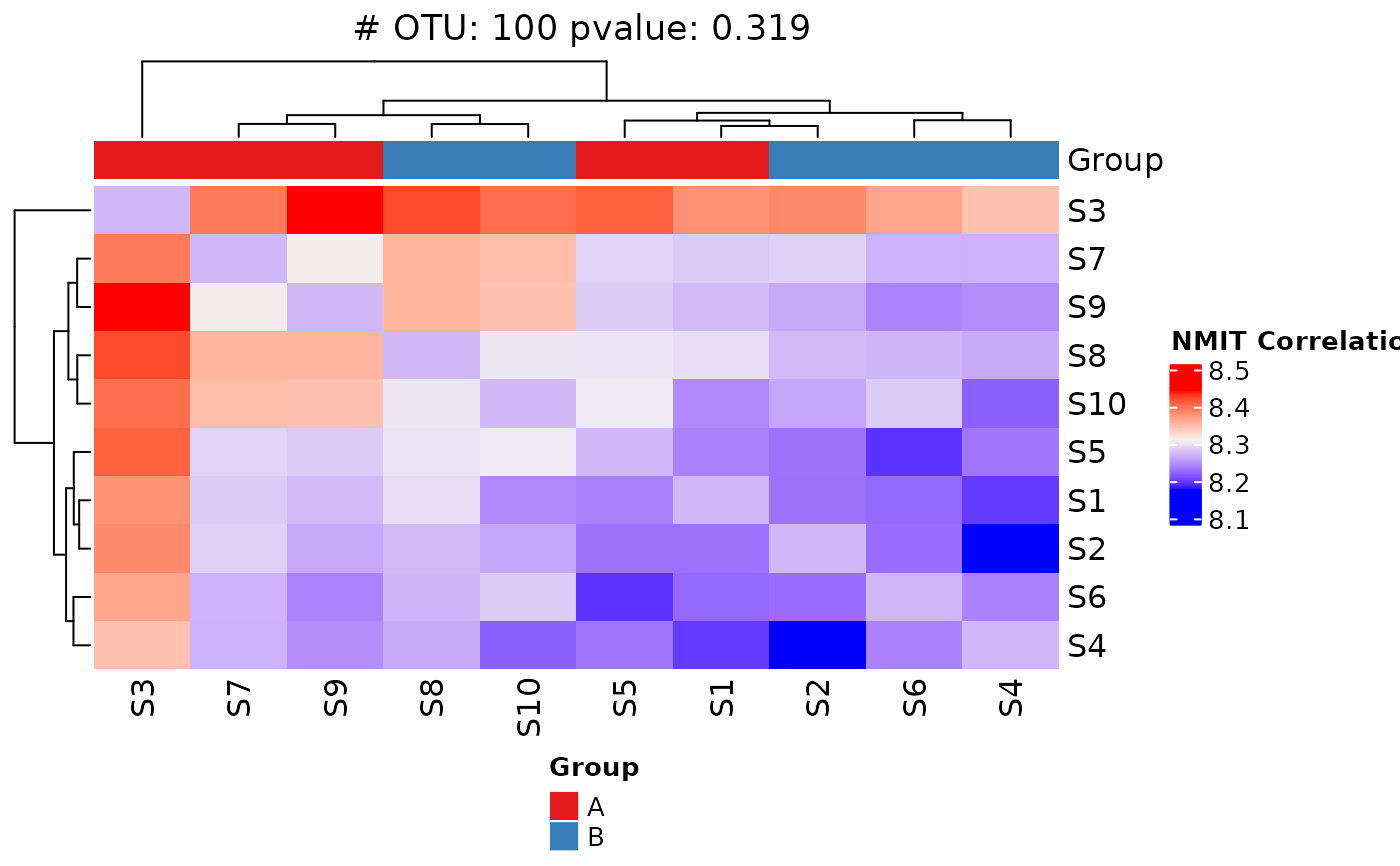
"F". *"M"or"m"specifies the maximum modulus of all the elements inx; *"O","o"or"1"specifies the one norm, (maximum absolute column sum); *"I"or"i"specifies the infinity norm (maximum absolute row sum); *"F"or"f"specifies the Frobenius norm (the Euclidean norm ofxtreated as if it were a vector)- heatmap
A logical value indicating whether to draw heatmap. The default is
TRUE.- classify
A logical value indicating whether to draw a classifier tree. The default is
FALSE.- fill_na
A number between 0 and 1 to fill
NAvalues. The default value is 0.- ...
Additional arguments to be passed to
ComplexHeatmap::Heatmap().
Value
This function returns an analysis of variance (ANOVA) table showing sources of variation, degrees of freedom, sequential sums of squares, mean squares, F statistics, partial R-squared and P values, based on 999 permutations.
Examples
dat <- system.file("extdata/MAE_small.RDS", package = "LegATo") |> readRDS()
NMIT(dat, unit_var = "Subject", fixed_cov = "Group", covariate_time = "Month")
 #> NULL
#> NULL